Stephen, Damjan & Allan co-author UK Biobank paper
Genome-Wide Genetic Data on ~500,000 UK Biobank Participants
Genome-wide genetic data on ~500,000 UK Biobank participants
Clare Bycroft, Colin Freeman, Desislava Petkova, Gavin Band, Lloyd T Elliott, Kevin Sharp, Allan Motyer, Damjan Vukcevic, Olivier Delaneau, Jared O'Connell, Adrian Cortes, Samantha Welsh, Gil McVean, Stephen Leslie, Peter Donnelly, Jonathan Marchini. doi: https://doi.org/10.1101/166298
Stephen Leslie, Damjan Vukcevic and Allan Motyer played a key role in the main data release for the UK Biobank, one of the most important resources for genetic medical research ever produced (http://www.ukbiobank.ac.uk/). The resource contains electronic medical records, phenotype data and genomic data for over 500,000 participants. Working as part of a small group of researchers (mostly at Oxford), the MIG researchers produced and validated the genetic data for the whole resource. Specifically, the MIG team produced and validated HLA types for the entire cohort. The associated paper is currently available on BioRxiv and will be published soon.
The preprint has already garnered considerable attention https://biorxiv.altmetric.com/details/22244577
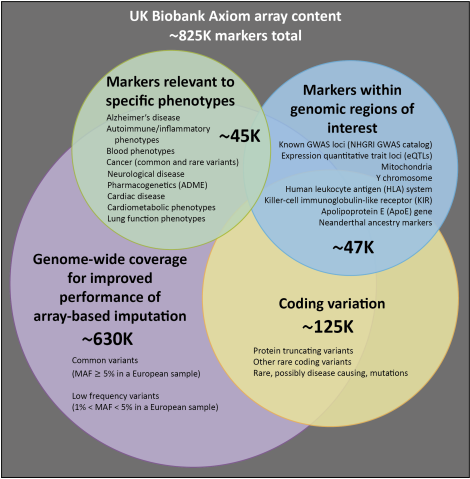
They describe the genome-wide genotype data (~805,000 markers) collected on all individuals in the cohort and its quality control procedures. Genotype data on this scale offers novel opportunities for assessing quality issues, although the wide range of ancestries of the individuals in the cohort also creates particular challenges. They also conducted a set of analyses that reveal properties of the genetic data (such as population structure and relatedness) that can be important for downstream analyses.
In addition, they phased and imputed genotypes into the dataset, using computationally efficient methods combined with the Haplotype Reference Consortium (HRC) and UK10K haplotype resource. This increases the number of testable variants by over 100-fold to ~96 million variants. They also imputed classical allelic variation at 11 human leukocyte antigen (HLA) genes, and as a quality control check of this imputation, they replicate signals of known associations between HLA alleles and many common diseases.
They describe tools that allow efficient genome-wide association studies (GWAS) of multiple traits and fast phenome-wide association studies (PheWAS), which work together with a new compressed file format that has been used to distribute the dataset. As a further check of the genotyped and imputed datasets, they performed a test-case genome-wide association scan on a well-studied human trait, standing height.
Figure 1: Summary of UK Biobank genotyping array content. This is a schematic representation of the different categories of content on the UK Biobank Axiom array. Numbers indicate the approximate count of markers within each category, ignoring any overlap.